
Testing, Confidence Intervals, Sample Size and Power for Comparing Two Binomial Rates
Source:R/gsBinomial.R, R/varBinomial.R
varBinomial.RdSupport is provided for sample size estimation, power, testing, confidence intervals and simulation for fixed sample size trials (that is, not group sequential or adaptive) with two arms and binary outcomes. Both superiority and non-inferiority trials are considered. While all routines default to comparisons of risk-difference, options to base computations on risk-ratio and odds-ratio are also included.
nBinomial() computes sample size or power using the method of
Farrington and Manning (1990) for a trial to test the difference between two
binomial event rates. The routine can be used for a test of superiority or
non-inferiority. For a design that tests for superiority nBinomial()
is consistent with the method of Fleiss, Tytun, and Ury (but without the
continuity correction) to test for differences between event rates. This
routine is consistent with the Hmisc package routines bsamsize and
bpower for superiority designs. Vector arguments allow computing
sample sizes for multiple scenarios for comparative purposes.
testBinomial() computes a Z- or Chi-square-statistic that compares
two binomial event rates using the method of Miettinen and Nurminen (1980).
This can be used for superiority or non-inferiority testing. Vector
arguments allow easy incorporation into simulation routines for fixed, group
sequential and adaptive designs.
ciBinomial() computes confidence intervals for 1) the difference
between two rates, 2) the risk-ratio for two rates or 3) the odds-ratio for
two rates. This procedure provides inference that is consistent with
testBinomial() in that the confidence intervals are produced by
inverting the testing procedures in testBinomial(). The Type I error
alpha input to ciBinomial is always interpreted as 2-sided.
simBinomial() performs simulations to estimate the power for a
Miettinen and Nurminen (1985) test comparing two binomial rates for
superiority or non-inferiority. As noted in documentation for
bpower.sim() in the HMisc package, by using testBinomial() you
can see that the formulas without any continuity correction are quite
accurate. In fact, Type I error for a continuity-corrected test is
significantly lower (Gordon and Watson, 1996) than the nominal rate. Thus,
as a default no continuity corrections are performed.
varBinomial computes blinded estimates of the variance of the
estimate of 1) event rate differences, 2) logarithm of the risk ratio, or 3)
logarithm of the odds ratio. This is intended for blinded sample size
re-estimation for comparative trials with a binary outcome.
Testing is 2-sided when a Chi-square statistic is used and 1-sided when a Z-statistic is used. Thus, these 2 options will produce substantially different results, in general. For non-inferiority, 1-sided testing is appropriate.
You may wish to round sample sizes up using ceiling().
Farrington and Manning (1990) begin with event rates p1 and p2
under the alternative hypothesis and a difference between these rates under
the null hypothesis, delta0. From these values, actual rates under
the null hypothesis are computed, which are labeled p10 and
p20 when outtype=3. The rates p1 and p2 are used
to compute a variance for a Z-test comparing rates under the alternative
hypothesis, while p10 and p20 are used under the null
hypothesis. This computational method is also used to estimate variances in
varBinomial() based on the overall event rate observed and the input
treatment difference specified in delta0.
Sample size with scale="Difference" produces an error if
p1-p2=delta0. Normally, the alternative hypothesis under
consideration would be p1-p2-delta0$>0$. However, the alternative can
have p1-p2-delta0$<0$.
Usage
ciBinomial(x1, x2, n1, n2, alpha = 0.05, adj = 0, scale = "Difference")
nBinomial(
p1,
p2,
alpha = 0.025,
beta = 0.1,
delta0 = 0,
ratio = 1,
sided = 1,
outtype = 1,
scale = "Difference",
n = NULL
)
simBinomial(
p1,
p2,
n1,
n2,
delta0 = 0,
nsim = 10000,
chisq = 0,
adj = 0,
scale = "Difference"
)
testBinomial(
x1,
x2,
n1,
n2,
delta0 = 0,
chisq = 0,
adj = 0,
scale = "Difference",
tol = 1e-11
)
varBinomial(x, n, delta0 = 0, ratio = 1, scale = "Difference")Arguments
- x1
Number of “successes” in the control group
- x2
Number of “successes” in the experimental group
- n1
Number of observations in the control group
- n2
Number of observations in the experimental group
- alpha
type I error; see
sidedbelow to distinguish between 1- and 2-sided tests- adj
With
adj=1, the standard variance with a continuity correction is used for a Miettinen and Nurminen test statistic This includes a factor of \(n / (n - 1)\) where \(n\) is the total sample size. Ifadjis not 1, this factor is not applied. The default isadj=0since nominal Type I error is generally conservative withadj=1(Gordon and Watson, 1996).- scale
“Difference”, “RR”, “OR”; see the
scaleparameter documentation above and Details. This is a scalar argument.- p1
event rate in group 1 under the alternative hypothesis
- p2
event rate in group 2 under the alternative hypothesis
- beta
type II error
- delta0
A value of 0 (the default) always represents no difference between treatment groups under the null hypothesis.
delta0is interpreted differently depending on the value of the parameterscale. Ifscale="Difference"(the default),delta0is the difference in event rates under the null hypothesis (p10 - p20). Ifscale="RR",delta0is the logarithm of the relative risk of event rates (p10 / p20) under the null hypothesis. Ifscale="LNOR",delta0is the difference in natural logarithm of the odds-ratio under the null hypothesislog(p10 / (1 - p10)) - log(p20 / (1 - p20)).- ratio
sample size ratio for group 2 divided by group 1
- sided
2 for 2-sided test, 1 for 1-sided test
- outtype
nBinomialonly; 1 (default) returns total sample size; 2 returns a data frame with sample size for each group (n1, n2; ifnis not input asNULL, power is returned inPower; 3 returns a data frame with total sample size (n), sample size in each group (n1, n2), Type I error (alpha), 1 or 2 (sided, as input), Type II error (beta), power (Power), null and alternate hypothesis standard deviations (sigma0, sigma1), input event rates (p1, p2), null hypothesis difference in treatment group means (delta0) and null hypothesis event rates (p10, p20).- n
If power is to be computed in
nBinomial(), input total trial sample size inn; this may be a vector. This is also the sample size invarBinomial, in which case the argument must be a scalar.- nsim
The number of simulations to be performed in
simBinomial()- chisq
An indicator of whether or not a chi-square (as opposed to Z) statistic is to be computed. If
delta0=0(default), the difference in event rates divided by its standard error under the null hypothesis is used. Otherwise, a Miettinen and Nurminen chi-square statistic for a 2 x 2 table is used.- tol
Default should probably be used; this is used to deal with a rounding issue in interim calculations
- x
Number of “successes” in the combined control and experimental groups.
Value
testBinomial() and simBinomial() each return a vector
of either Chi-square or Z test statistics. These may be compared to an
appropriate cutoff point (e.g., qnorm(.975) for normal or
qchisq(.95,1) for chi-square).
ciBinomial() returns a data frame with 1 row with a confidence
interval; variable names are lower and upper.
varBinomial() returns a vector of (blinded) variance estimates of the
difference of event rates (scale="Difference"), logarithm of the
odds-ratio (scale="OR") or logarithm of the risk-ratio
(scale="RR").
With the default outtype=1, nBinomial() returns a vector of
total sample sizes is returned. With outtype=2, nBinomial()
returns a data frame containing two vectors n1 and n2
containing sample sizes for groups 1 and 2, respectively; if n is
input, this option also returns the power in a third vector, Power.
With outtype=3, nBinomial() returns a data frame with the
following columns:
- n
A vector with total samples size required for each event rate comparison specified
- n1
A vector of sample sizes for group 1 for each event rate comparison specified
- n2
A vector of sample sizes for group 2 for each event rate comparison specified
- alpha
As input
- sided
As input
- beta
As input; if
nis input, this is computed- Power
If
n=NULLon input, this is1-beta; otherwise, the power is computed for each sample size input- sigma0
A vector containing the standard deviation of the treatment effect difference under the null hypothesis times
sqrt(n)whenscale="Difference"orscale="OR"; whenscale="RR", this is the standard deviation timesqrt(n)for the numerator of the Farrington-Manning test statisticx1-exp(delta0)*x2.- sigma1
A vector containing the values as
sigma0, in this case estimated under the alternative hypothesis.- p1
As input
- p2
As input
- p10
group 1 event rate used for null hypothesis
- p20
group 2 event rate used for null hypothesis
References
Farrington, CP and Manning, G (1990), Test statistics and sample size formulae for comparative binomial trials with null hypothesis of non-zero risk difference or non-unity relative risk. Statistics in Medicine; 9: 1447-1454.
Fleiss, JL, Tytun, A and Ury (1980), A simple approximation for calculating sample sizes for comparing independent proportions. Biometrics;36:343-346.
Gordon, I and Watson R (1985), The myth of continuity-corrected sample size formulae. Biometrics; 52: 71-76.
Miettinen, O and Nurminen, M (1985), Comparative analysis of two rates. Statistics in Medicine; 4 : 213-226.
Author
Keaven Anderson keaven_anderson@merck.com
Examples
# Compute z-test test statistic comparing 39/500 to 13/500
# use continuity correction in variance
x <- testBinomial(x1 = 39, x2 = 13, n1 = 500, n2 = 500, adj = 1)
x
#> [1] 3.701266
pnorm(x, lower.tail = FALSE)
#> [1] 0.0001072634
# Compute with unadjusted variance
x0 <- testBinomial(x1 = 39, x2 = 23, n1 = 500, n2 = 500)
x0
#> [1] 2.098083
pnorm(x0, lower.tail = FALSE)
#> [1] 0.0179489
# Perform 50k simulations to test validity of the above
# asymptotic p-values
# (you may want to perform more to reduce standard error of estimate)
sum(as.double(x0) <=
simBinomial(p1 = .078, p2 = .078, n1 = 500, n2 = 500, nsim = 10000)) / 10000
#> [1] 0.017
sum(as.double(x0) <=
simBinomial(p1 = .052, p2 = .052, n1 = 500, n2 = 500, nsim = 10000)) / 10000
#> [1] 0.0188
# Perform a non-inferiority test to see if p2=400 / 500 is within 5% of
# p1=410 / 500 use a z-statistic with unadjusted variance
x <- testBinomial(x1 = 410, x2 = 400, n1 = 500, n2 = 500, delta0 = -.05)
x
#> [1] 2.807617
pnorm(x, lower.tail = FALSE)
#> [1] 0.002495478
# since chi-square tests equivalence (a 2-sided test) rather than
# non-inferiority (a 1-sided test),
# the result is quite different
pchisq(testBinomial(
x1 = 410, x2 = 400, n1 = 500, n2 = 500, delta0 = -.05,
chisq = 1, adj = 1
), 1, lower.tail = FALSE)
#> [1] 0.005012758
# now simulate the z-statistic witthout continuity corrected variance
sum(qnorm(.975) <=
simBinomial(p1 = .8, p2 = .8, n1 = 500, n2 = 500, nsim = 100000)) / 100000
#> [1] 0.02465
# compute a sample size to show non-inferiority
# with 5% margin, 90% power
nBinomial(p1 = .2, p2 = .2, delta0 = .05, alpha = .025, sided = 1, beta = .1)
#> [1] 2697.607
# assuming a slight advantage in the experimental group lowers
# sample size requirement
nBinomial(p1 = .2, p2 = .19, delta0 = .05, alpha = .025, sided = 1, beta = .1)
#> [1] 4131.9
# compute a sample size for comparing 15% vs 10% event rates
# with 1 to 2 randomization
nBinomial(p1 = .15, p2 = .1, beta = .2, ratio = 2, alpha = .05)
#> [1] 1191.041
# now look at total sample size using 1-1 randomization
n <- nBinomial(p1 = .15, p2 = .1, beta = .2, alpha = .05)
n
#> [1] 1079.853
# check if inputing sample size returns the desired power
nBinomial(p1 = .15, p2 = .1, beta = .2, alpha = .05, n = n)
#> [1] 0.8
# re-do with alternate output types
nBinomial(p1 = .15, p2 = .1, beta = .2, alpha = .05, outtype = 2)
#> n1 n2
#> 1 539.9264 539.9264
nBinomial(p1 = .15, p2 = .1, beta = .2, alpha = .05, outtype = 3)
#> n n1 n2 alpha sided beta Power sigma0 sigma1 p1
#> 1 1079.853 539.9264 539.9264 0.05 1 0.2 0.8 0.6614378 0.6595453 0.15
#> p2 delta0 p10 p20
#> 1 0.1 0 0.125 0.125
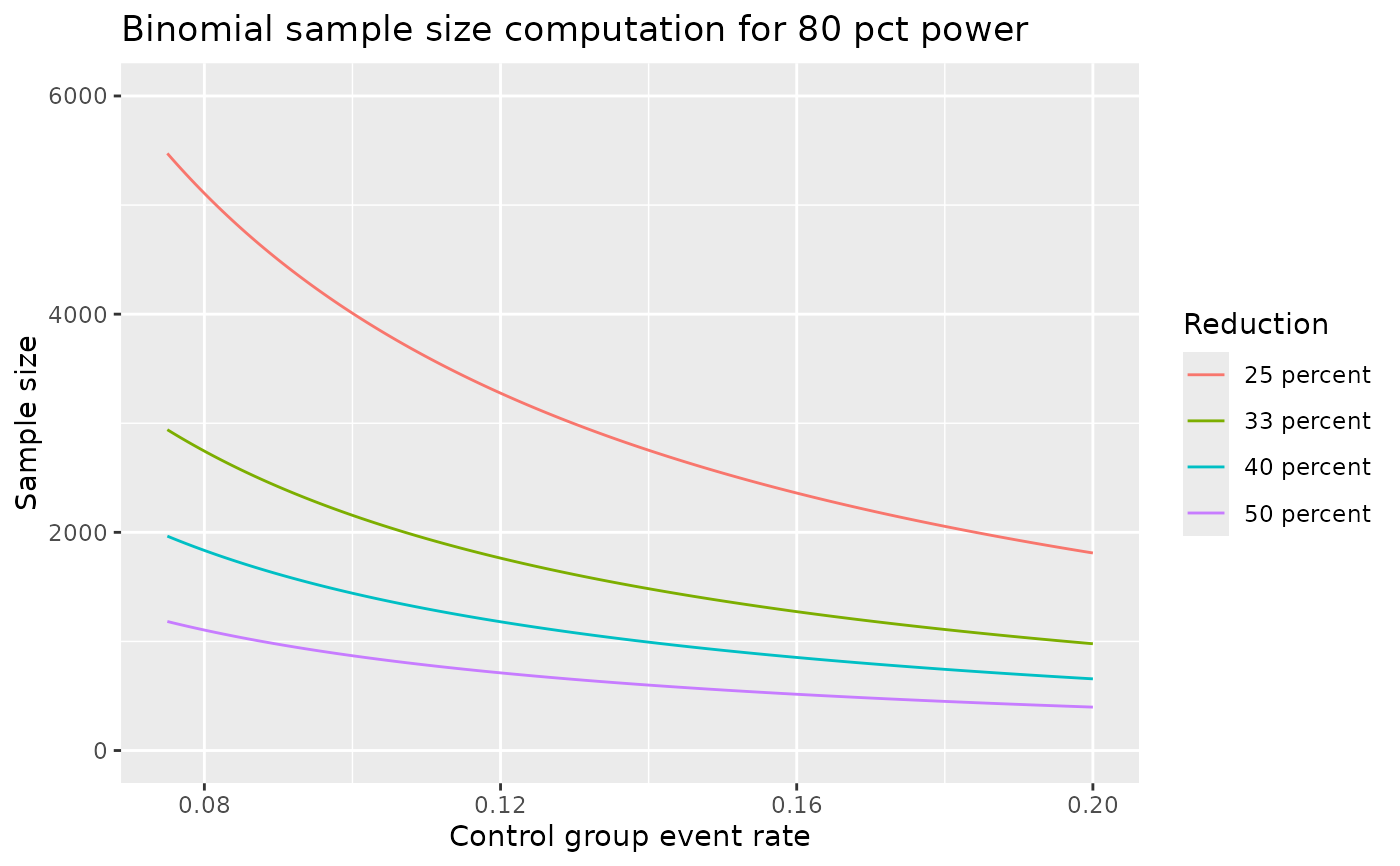
# look at power plot under different control event rate and
# relative risk reductions
library(dplyr)
#>
#> Attaching package: ‘dplyr’
#> The following objects are masked from ‘package:stats’:
#>
#> filter, lag
#> The following objects are masked from ‘package:base’:
#>
#> intersect, setdiff, setequal, union
library(ggplot2)
p1 <- seq(.075, .2, .000625)
len <- length(p1)
p2 <- c(p1 * .75, p1 * 2/3, p1 * .6, p1 * .5)
Reduction <- c(rep("25 percent", len), rep("33 percent", len),
rep("40 percent", len), rep("50 percent", len))
df <- tibble(p1 = rep(p1, 4), p2, Reduction) |>
mutate(`Sample size` = nBinomial(p1, p2, beta = .2, alpha = .025, sided = 1))
ggplot(df, aes(x = p1, y = `Sample size`, col = Reduction)) +
geom_line() +
xlab("Control group event rate") +
ylim(0,6000) +
ggtitle("Binomial sample size computation for 80 pct power")
 # compute blinded estimate of treatment effect difference
x1 <- rbinom(n = 1, size = 100, p = .2)
x2 <- rbinom(n = 1, size = 200, p = .1)
# blinded estimate of risk difference variance
varBinomial(x = x1 + x2, n = 300, ratio = 2, delta0 = 0)
#> [1] 0.001269333
# blinded estimate of log-risk-ratio variance
varBinomial(x = x1 + x2, n = 300, ratio = 2, delta0 = 0, scale = "RR")
#> [1] 0.1457143
# blinded estimate of log-odds-ratio variance
varBinomial(x = x1 + x2, n = 300, ratio = 2, delta0 = 0, scale = "OR")
#> [1] 0.1772584
# compute blinded estimate of treatment effect difference
x1 <- rbinom(n = 1, size = 100, p = .2)
x2 <- rbinom(n = 1, size = 200, p = .1)
# blinded estimate of risk difference variance
varBinomial(x = x1 + x2, n = 300, ratio = 2, delta0 = 0)
#> [1] 0.001269333
# blinded estimate of log-risk-ratio variance
varBinomial(x = x1 + x2, n = 300, ratio = 2, delta0 = 0, scale = "RR")
#> [1] 0.1457143
# blinded estimate of log-odds-ratio variance
varBinomial(x = x1 + x2, n = 300, ratio = 2, delta0 = 0, scale = "OR")
#> [1] 0.1772584